The term “undruggable” has entered common industry vernacular over the past two decades to describe biologic targets that couldn’t be addressed via narrow conventional pharmacologies involving catalytic-site inhibitors and antibodies.
The “druggable genome” historically included intracellular catalytic proteins, (e.g., enzymes like kinases) and extracellular or transmembrane targets (e.g., ion channels, receptors, secreted proteins), representing only a small fraction of the human proteome – maybe 10 percent. But new therapeutic modalities have emerged that offer the potential to open the rest of the proteome to exploration, including targets like transcription factors, scaffolds in protein complexes, and signaling adaptor molecules.
Targeted protein degradation is one of the most compelling of these new modalities: this approach works by creating heterobifunctional chemistries which are able to simultaneously bind to proteins inside of cells and tag them for degradation by hijacking the cell’s normal protein disposal machinery, the ubiquitin-proteasome system. It’s an elegant, catalytic mechanism creating post-translational, chemically-induced knockdown of disease-causing proteins, and thus offers enormous potential to address previously intractable targets.
 Today we’ve announced the launch of a new company, Kymera Therapeutics, which aims to create a world-leading therapeutics platform in this emerging field, discovering and developing novel first-in-class medicines that offer unique degradation-induced pharmacology that we believe will result in clinically differentiated efficacy.
Today we’ve announced the launch of a new company, Kymera Therapeutics, which aims to create a world-leading therapeutics platform in this emerging field, discovering and developing novel first-in-class medicines that offer unique degradation-induced pharmacology that we believe will result in clinically differentiated efficacy.
But before jumping into the Kymera story, it’s worth exploring the history of the field and what makes the modality so special – which forms the backdrop for what got us excited about this technology at Atlas.
Brief History of Targeted Protein Degradation
Most proteins are degraded through the proteasome as part of normal turnover; in fact, the ubiquitin-proteasome system is the primary conduit for the majority of protein turnover and removal of misfolded proteins in eukaryotic cells. They get tagged with ubiquitin, a small protein recognized by the cell’s trash process, via enzymes called E3 ligases. Once tagged, these proteins are unfolded and channeled into the proteasome, a multi-subunit catalytic machine that is effectively the garbage disposal of the cell, cutting proteins into smaller peptide fragments. The rules of this process have been the subject of lots of exploration since the 1980s (like one of my favorites, Varshavsky’s N-end rule). By coincidence, several chapters of my graduate thesis in the 1990s described how the proteasome degrades antigenic proteins and helps process them for presentation to the immune system. Because of that connection, I’ve always loved the elegance of the proteasome system, and perhaps that’s why this modality has been of great interest to me for years.
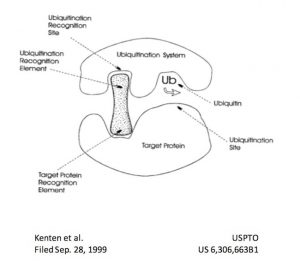 As the rules governing protein homeostasis became increasingly clear, the concept of hijacking the system to exploit targeted protein degradation emerged. The first known reference to the concept of this new modality can be found in a now abandoned patent filing by John Kenten et al in 1999, which describes the concept of a heterobifunctional molecule that engages a target and an E3 ligase to trigger the target’s ubiquitination and subsequent degradation. The cover illustration of the patent (right) is amazingly prescient, effectively describing the essence of the modality 15 years before the field would show in vivo proof of concept for targeted protein degradation.
As the rules governing protein homeostasis became increasingly clear, the concept of hijacking the system to exploit targeted protein degradation emerged. The first known reference to the concept of this new modality can be found in a now abandoned patent filing by John Kenten et al in 1999, which describes the concept of a heterobifunctional molecule that engages a target and an E3 ligase to trigger the target’s ubiquitination and subsequent degradation. The cover illustration of the patent (right) is amazingly prescient, effectively describing the essence of the modality 15 years before the field would show in vivo proof of concept for targeted protein degradation.
Academically, the field began to take off in the early 2000s. Pioneers of the field of targeted protein degradation over the last seventeen years include Ray Deshaies of Caltech (now Amgen), Craig Crews of Yale, and Jay Bradner of DFCI (now Novartis), among many others. Early demonstrations included peptidic degrader chimeras for Skp1 (Sakamoto et al 2001) and small molecule degraders against MDM2 (Schneekloth et al 2008). Critical in-vivo demonstration of small molecule degraders came in 2015 from both the Bradner lab on BRD4 (Winter et al 2015) and Crews lab on ERR and RIPK2 (Bondeson et al 2015). These and other key papers have firmly established the modality as “ready for prime time” in terms of therapeutic exploration. For those interested, a new and thorough review of the modality was just published in Annual Reports of Medicinal Chemistry, written by Kymera co-founder Nello Mainolfi.
Unique Therapeutic Profiles
The pharmacology of these targeted degraders is truly distinctive from traditional active-site inhibitors. In most cases, the latter requires binding to a protein’s active site where it competes against endogenous ligands; its activity depends on an equilibrium related to how tightly it binds (affinities) and its concentration in the body. If the drug isn’t around at sufficiently high levels, there’s not going to be any activity. Often the drug needs to stay above a certain threshold (like the IC90) for any functional inhibition to occur. This is the conventional “PK–PD relationship.”
But degraders change all that. They don’t need to bind active sites – they can bind anywhere. This flexibility, along with spatial parameters of the target-ligase interactions, affords greater selectivity against other proteins. And once a degrader binds and recruits a ligase, it triggers an irreversible process of protein degradation, eliminating the target protein from the cell. The approach is catalytic in nature, meaning the drugs can keep on working until they can’t find more protein to degrade. What limits their effect is the re-synthesis of new target proteins (usually 24-72 hours), and the tissue-level PK and metabolic stability of the drug itself in cells. But fundamentally there can be a massive disconnect between PK and PD with this approach, creating a totally new way to think about targeting proteins via this chemical knockdown strategy. These features could open up new avenues for achieving profound efficacy, improved safety, less frequent dosing and compliance benefits, as well as potentially enabling new route of administration and combination opportunities. This is truly exciting for patients and for our prospects of advancing the standard of care.
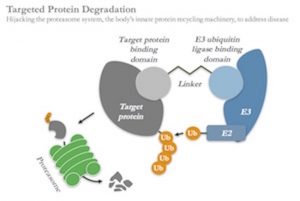 Further, because degraders don’t need active enzymatic sites, they can bind to and degrade a huge range of previously intractable proteins such as adaptor molecules, transcription factors, pseudo-enzymes, and scaffolding proteins. Kymera has already demonstrated in-vitro proof of concept for several of these different classes of proteins. This allows us to systemically and completely “mine” a particular pathway by targeting proteins up and down the signaling process for degradation, from cell surface receptors to transcription factors.
Further, because degraders don’t need active enzymatic sites, they can bind to and degrade a huge range of previously intractable proteins such as adaptor molecules, transcription factors, pseudo-enzymes, and scaffolding proteins. Kymera has already demonstrated in-vitro proof of concept for several of these different classes of proteins. This allows us to systemically and completely “mine” a particular pathway by targeting proteins up and down the signaling process for degradation, from cell surface receptors to transcription factors.
Moving Into Mainstream
With the evolution of the scientific basis of this new modality over the past decade, several startups and many pharma companies have also begun pursuing it. Craig Crews co-founded Arvinas, which raised its first round of funding in 2013; it’s a great story and we’re rooting for the clinical success of their advancing androgen and estrogen programs. Jay Bradner co-founded C4 Therapeutics with some Dana Farber colleagues in the fall of 2015 before leaving for Novartis; C4 is another solid biotech in the field doing high quality work. Many major pharma companies have efforts in the space, including Celgene, Takeda, GSK, Novartis and AZ among others.
Along with the proliferation of companies engaged in the space is the proliferation of names: PROTACs, Degronimids, SNIPERs, etc… Fundamentally, these are all riffs on the same targeted protein degradation theme as described by Kenten et al in 1999: heterobifunctional molecules that promote degradation by bringing targets in proximity to ubiquitin ligases.
I’m a firm believer that with new modalities like this, a rising tide lifts all the boats. In the early days of antibodies, a number of players explored different technical solutions to similar problems – and today we’ve got over 55 antibodies approved by the FDA, discovered across a whole range of underlying techniques and approaches (recent blog on topic here). I’m convinced targeted protein degradation will become one of many successful modalities that advance over the next decade, with multiple strong players in the space.
Since others are active in the field, we frequently get the question, what is our point of differentiation versus the “competition.” My first response is always that this is a broad new modality with plenty of freedom to operate across potentially every therapeutic area and disease state. There will be many players pursuing this great modality to bring innovative therapies to patients. New efforts in pharma, and in startups, will happen. In the long arc of biotechnology history, winners are those that bring real medicines to patients, which involves creating new therapies targeting specific disease-causing proteins and then translating them to the clinic. Platforms must become products. With 90% of the proteome to now play with, there will be plenty of room to build great companies here around different product opportunities.
But there are two points of real differentiation to highlight.
First, we spent the last eighteen months in stealth mode building an integrated drug discovery platform that takes a novel, systematic approach to understanding the modality’s unique ternary complex formation (drug + target + ligase) and how to optimize its catalytic potential. This incorporates a systematic patient/disease-centric target selection process powered by a proprietary mathematical, data-driven prediction model which helps us pick the ideal target and target-ligase pairs to engage in order to drastically increase probability of success. Our patent strategy, which also includes novel ligands to E-3 ligases, protects these insights. We’ve also brought full-scale proteomics into the early discovery funnel to understand degradation across thousands of proteins, amongst other key parameters. We’re exploring a diverse set of methods to expand the modality’s reach across multiple routes of administration, including oral delivery.
Second, we’re mining novel pathways with this modality. Instead of going after single targets like hormone receptors and bromodomains (like some of the effort at Arvinas and C4), we’ve been initially focused on exploring broader pathways in oncology, immunology, and fibrosis. For example, take toll-like receptor and interleukin-1 signaling through the Myddosome (e.g., TLRs, IL1/18/33R, TRAFs, MyD88, IRAK4, IRAK1, the negative regulator IRAK3); we have emerging in-vivo and in-vitro data on degraders addressing several of these targets, and are advancing into lead optimization. In line with our investment thesis, these early leads appear to show significantly differentiated pharmacology relative to small molecule inhibitors of proteins in the pathway.
Birth of Kymera Therapeutics
Every good biotech has its creation story, and Kymera is no different. Inspired by the growing body of academic evidence around the in-vivo potential of this modality, we committed to creating a targeted protein degradation startup in early 2016. Two serial Atlas entrepreneurs, Nello Mainolfi (former head of drug discovery for Raze Therapeutics, ex-Novartis) and Stuart Chaffee (former EIR with Magenta, ex-Biogen), worked with me to co-found the company, along with Atlas associate Steve Robinette. In addition, Kymera has recruited an exceptional broader team that bring prior tours of duty at Novartis, Biogen, Moderna, Arvinas, and beyond.
Atlas seeded the company in the spring of 2016 and have incubated Kymera with us since. To access lab space, we set up a “second site” no more than a hundred yards down Main Street at LabCentral, leveraging a “Golden Ticket” from J&J/JLABS. This dual footprint model, while not always ideal, has worked very well for Kymera in its infancy and allowed us to achieve an incredible amount of progress in a capital efficient manner.
During our six quarters in stealth seed phase, we achieved all of our initial goals: replicating the academic data, in vitro confirmation of cellular degradation of several targets in cultured lines and primary cells, discovery of novel E3 ligase ligands, identification of ligands to traditionally undruggable proteins and in-vivo proof of principle against targets of interest.
With these data in hand, and momentum around the emerging pipeline in oncology, immuno-oncology, inflammation, and fibrosis, we raised a $30M Series A round along with Lilly Ventures and Amgen Ventures. A trusted co-investor with us at Nimbus and Lysosomal Therapeutics, and skilled medicinal chemist, Lilly Ventures’ Steve Hall joined the Board. Amgen Ventures invested in part because of Ray Deshaies’ commitment to the space and his pre-Amgen advisory work with the founding team at Kymera in its early days.
As part of this Series A, it was time for me to move on as the founding CEO, and we were fortunate to recruit Laurent Audoly, a seasoned Pharma R&D executive, to join us. Laurent was previously Head of R&D at Pierre Fabre, and before that was CSO at Pieris – a novel modality company in Europe. He had more than a dozen years at Merck and Pfizer early in his career, and had worked closely with members of the Atlas family, like Nimbus’ Don Nicholson while at Merck. The stars aligned in the middle of the summer when the gravity of Cambridge’s biotech ecosystem, and the lure of Kymera, was able to pull Laurent and his family out of Toulouse, France. He joined us in August and has hit the ground running.
In parallel to Kymera’s scientific progress, in recent months we’ve initiated partnering conversations (as most young platform companies do). We shifted the corporate structure of Kymera into the Nimbus-like LLC structure, enabling a broader range of potential future deals with Pharma in a tax-efficient manner.
We have assembled a great group of academic advisors: heterobifunctional chemistry and modeling expert David Spiegel at Yale; pioneer in deep proteomics Steve Carr at the Broad Institute; and, world renowned scientists and experts in E3 ligase biology Michele Pagano at NYU and Ning Zheng from University of Washington, both also HHMI. In addition, Kymera is building a strong network of program-specific advisors in disease and translational biology that will be announced in due course.
It’s always an exciting time launching a new company into the world, and Kymera is no exception.
We’re aiming to drug the undruggable – and bring first-in-class medicines to patients that harness the power of targeted protein degradation to deliver unique therapeutic benefits. Onward and upward.