Extreme clinical phenotypes caused by rare genetic variations have been the basis of some of the most exciting drug targets in today’s pharmaceutical pipeline: PCSK9 in cholesterol, sclerostin in bone diseases, Nav1.7 for pain. The rare gene signatures underlying these programs can provide a big boost to the confidence in rationale (CIR) for working on novel drug approaches, especially those that have no clinical precedent for pharmacologic intervention.
Target selection is one of the biggest drivers of risk for a young discovery-stage startup or a similarly-staged project team in Pharma. Numerous analyses in the past show a striking difference in attrition with clinically precedented vs novel targets; in short, chasing targets with best-in-class profiles has historically been more successful than first-in-class novel approaches. But in today’s world, where the demand and requirement for more transformative innovation is much higher than in the past, understanding how to better prosecute those novel targets has become critical.
This is where understanding extreme phenotypes and the impact of “Nature’s Experiments” can greatly help on target prioritization and interrogation; since target validation is a hugely important risk driver, and any insight enabling one to increase, or decrease, CIR can be of real value.
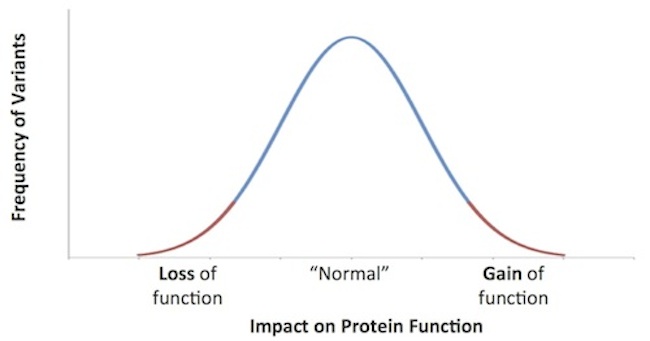
There are obviously different types of signals in genetics. Broadly speaking, variations like SNPs or deletions can lead to either gain of function or loss of function impacts (e.g., more/less signaling, enhanced/repressed expression, increased/decreased enzyme activity). The most profound gains or losses in function are the rare and most instructive phenotypes, and these have been the subject of much discussion (see this great paper by Sasha Kamb, Sean Harper, and Kari Stefansson at Amgen/DeCode, as well as this David Shaywitz piece).
Another excellent paper last fall from Robert Plenge, Ed Scolnick, and David Altshuler at the Broad Institute, explores the topic further, including “dose response” curves derived from Nature’s Experiments emphasizing its usefulness as tool to pick and prioritize targets. The authors explore the use of a ‘genetic therapeutic index’ of specific targets – where Nature has shown both the efficacy and safety effect of knocking out or augmenting a protein’s activity. For those interested in a thorough discussion of the topic, both of the above papers are well worth reading.
Although common variants and their cumulative impact on disease risk and health are important, the rare or less frequent tails of this gain/loss of function distribution are where drug hunters searching for CIR insight are focused.
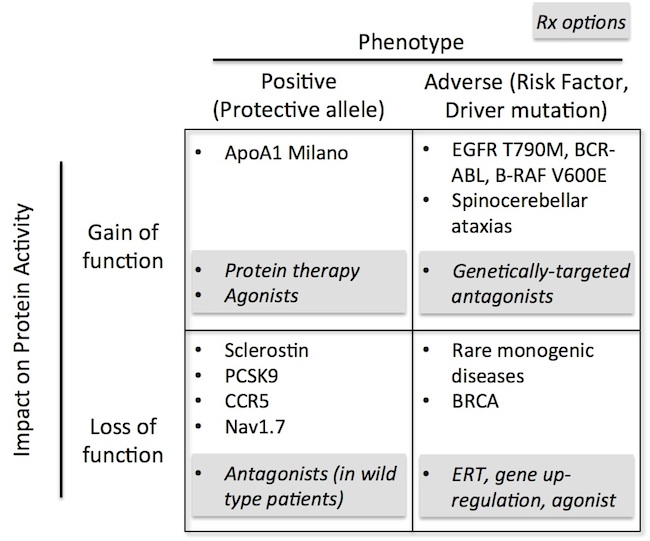
 These alterations in physiologic activity, when compared to “normal” individuals, can manifest as either protective, positive phenotypes (improved health) or as adverse phenotypes (e.g., risk factors or disease drivers). The chart below illustrates these differences, and highlights some of the well-known examples in medicine today. Each of these quadrants obviously has different implications for how to think about the therapeutic opportunity.
These alterations in physiologic activity, when compared to “normal” individuals, can manifest as either protective, positive phenotypes (improved health) or as adverse phenotypes (e.g., risk factors or disease drivers). The chart below illustrates these differences, and highlights some of the well-known examples in medicine today. Each of these quadrants obviously has different implications for how to think about the therapeutic opportunity.
Where a loss-of-function looks protective, like Nav1.7, Nature would suggest antagonizing those channels would be beneficial in “normal” individuals suffering from pain. Similarly, where a gain-of-function is driving a disease, like many cancer mutations, therapeutics to antagonize those functions would likely be beneficial; further, in that quadrant, the genetics can also enable patient stratification and selection.
Protective gain-of-function alleles, like ApoA-1 Milano in HDL elevation, would suggest protein supplementation or agonist strategies should work. Lastly, loss of function deletions, variants, or dominant negatives leading to adverse phenotypes, like most rare genetic diseases and some cancer risk factor genes (e.g, BRCA), would require therapeutic interventions bringing back gene function: enzyme replacement therapy, gene or mRNA therapy, augmenting expression of the “normal” copy of the gene, or increasing protein function.
Importantly, target validation can extend to the more general concept of “pathway validation”: rare variants up and down a signaling cascade can highlight phenotypic impact of interrogating a broader pathway of biology. For instance, blocking wnt family members involved with sclerostin in different bone disorders.
If approached correctly, leveraging these risk-reducing insights from nature to help prioritize “target selection” can help an early drug discovery program improve its odds of seeing a clinically meaningful outcome in 3-6 years. As we handicap the risk profiles of the 100s of drug discovery stage startup opportunities we see each year, being mindful of Nature’s experiments in genetics can help us get a better handle on the likelihood a target is worth pursuing.
Because we are a firm focused in large part on drug discovery, target selection is a critical variable to get right, and we apply these whatever insights we can glean from genetics into target prioritization across our portfolio. While not all of the ~60 or so drug programs active in our companies fall into the “tails” of the distribution above, a reasonable percentage have strong genetic evidence. Here are a dozen or so diverse examples meant to illustrate the breadth of the approach:
Gain of Function alleles with adverse phenotypes:
- MyD88-activating mutations creating IRAK4-dependency in liquid tumors such as DLBCL and Waldenstrom’s (ref): Nimbus Discovery
- Ryanodine mutations causing calcium dysregulation in CPVT patients, supporting RyR2 blockade (ref): Numerate
- Spinocerebellar ataxias (1,2,3,6,7) are CAG-repeat gain-of-function channelopathies, validating negative channel modulators (ref); Ataxion
- Different PAD4 gene variants causing mRNA stabilization and increased citrullination heighten risk of rheumatoid arthritis (ref): Padlock Therapeutics
- Tyk2-activating driver mutations in T-ALL, supporting a conventional “targeted therapy” approach in that leukemia (ref): Nimbus Discovery
Loss of function alleles with adverse phenotypes
- Glucocerebrosidase (GBA) mutations in Gauchers Disease, and as important risk factors for Parkinson’s disease (ref): Lysosomal Therapeutics Inc
- GPR120 loss-of-function allele linked to higher risk of obesity, supporting agonism of this lipid GPCR in metabolic disease (ref): Numerate
- b-myosin heavy chain mutations in hypertrophic cardiomyopathy, supporting strategies to up-regulate of alternative myosin isoforms (ref): Miragen Therapeutics
- TSC mutations in mTor pathway cause over activation and tumor/hamartoma syndromes, supporting selective inhibitors of mTor for both theses rare disease and common cancers (ref): Calorics
- Loss of function in frataxin leading to Friedreich’s Ataxia; upregulation of functional copy would be beneficial (ref): RaNA Therapeutics
Loss of function alleles with protective phenotypes:
- BH4 de novo synthesis pathway blockade linked to pain protection (GCH1), supporting of blocking the BH4 pathway in pain (ref): Quartet Medicine
- Tyk2 rare alleles showing protection against RA and lupus, supportive of Tyk2-selective inhibition in autoimmune disease (ref): Nimbus Discovery
- Atrial atriuretic peptide minor allele losing its 3’ UTR miR-425 binding site showing protection from hypertension, supportive of anti-miR–425 therapy in resistant hypertension (ref): Miragen Therapeutics
Gain of function genetics with beneficial phenotype
- Increased copy number of SMN2 correlates with less severe clinical outcomes in spinal muscular atrophy patients, supports that up-regulation of SMN2 expression would be beneficial (ref): RaNA Therapeutics
These types of targets are certainly not unique to Atlas; other early stage VCs invest in companies and targets with similar human genetic profiles. For example, Third Rock Venture’s portfolio has a number of targets validated through “tail” variations in gene function: Agios’ inborn error of metabolism program acround PKR, MyoKardia’s focus on myosin in hypertrophic cardiomyopathy, Global Blood’s aim to augment mutant hemoglobin’s oxygen binding, etc… Similar portfolios can presumably be found elsewhere in the early stage venture community.
Smart target selection is half the battle against project attrition, if not more. Although there are any number of ways for drug candidates to fail for reasons other than the target’s role in the disease, finding drug matter against a target is a more “manageable” risk than chasing irrelevant biology, or so says this former molecular biologist…