This blog was written by Jeb Keiper, CEO of Nimbus Therapeutics LLC, as part of the From The Trenches feature of LifeSciVC.
It is with great excitement that we at Nimbus are ringing in our 10th birthday this month. It’s been a momentous decade of ups and downs, many of them chronicled in dozens of posts here on LifeSciVC. At 10 years and still going, we’re proud to have reached a milestone that only about 10% of startups achieve.
We are using this 10th birthday as a time to look back on how well Nimbus has carried out its founding principles and to look ahead – as we define our vision for how the next decade of Nimbus should look. So, first, to revisit where we came from…
“Project Troubled Water”
The idea for Nimbus started kicking around in 2008 after Bruce Booth of Atlas Venture met Ramy Farid, CEO of Schrödinger. Schrödinger was just putting the finishing touches on its WaterMap™ software, which predicted the locations and free energies of water molecules in protein binding sites, providing a virtual map for designing molecules and therefore a hypothetical means to improve drug discovery. Booth and Farid worked on plans for “Project Troubled Water,” which was mercifully renamed Nimbus Discovery by the time the Series A rolled around. After two years incubating as a seed company, Nimbus came out of stealth in March 2011 and was highlighted in the blog post Discovering Nimbus. There were 3 fundamental founding hypotheses strung together in the creation of Nimbus — hypotheses that, 10 years on, we can now share some conclusions around.
Hypothesis 1: In silico drug discovery was ready for prime time
After 30 years of first hyped, then deflated expectations around computer applications in drug discovery, there were many eye-rolls on this one in Nimbus’ early years. The reasons to believe, however, were sound: a physics-based approach to calculate true free energies of ligands binding to proteins via an advanced force field (OPLS_2005 at the time) brought the differences in binding free energies down within 1 kcal per mole from in silico to experimental. Those quantum mechanics-based physics calculations thus became accurate enough to make some decent predictions. At the same time, the massive compute horsepower needed to run these calculations had come online: switching from CPUs to more powerful GPUs (Graphics Processing Units), readily available at the time in large part thanks to the video game industry, coupled with cloud computing access, provided enough “big compute power” to fuel the physics calculations in a way never before possible.
What was unique about Nimbus’ approach to using in silico methods was that by design there were no other paths to chemistry for the scientists at Nimbus. Working with our colleagues at Schrödinger was the only way to progress drug programs via software application. The going was tough too; the early years were at times frustrating and saw the failure of several programs. However, that failure was beautiful. The intense cooperation between the drug discovery experts at Nimbus and the computational experts at Schrödinger was the true battlefield testing, which led to new software creations like W-Score™. More importantly for Nimbus, the improvements ultimately led to success in developing quality chemical matter, including the first molecule ever prospectively designed by a computer to achieve first-in-human: NDI-010976, now known as Gilead’s firsocostat, a hepatotropic, allosteric inhibitor of acetyl CoA carboxylase, currently in Phase 2b studies for NASH.
Our next breakthrough, an exquisitely potent and selective allosteric Tyk2 modulator entering the clinic within the year, has achieved >100 fold improved selectivity for the target compared to BMS’ lauded Tyk2 program. While computational methods are important, they are still not everything. On one undisclosed target program, the computational modeling for months was unable to reliably predict selectivity. Here is where our high caliber, experienced chemists drove the program forward. The empirical approach still has a place in drug discovery, and as these designs bore fruit, the Schrödinger developers were able to modify the computational algorithms to the point where they reliably advanced the chemistry rapidly again: an exemplar of the teamwork between Nimbus and Schrödinger. So yes, we have certainly validated that cutting-edge in silico-driven discovery works, but only augmented with real AI, and by that, I mean “Actual Intelligence” – the talented drug hunters behind it.
Hypothesis 1: In silico drug discovery was ready for prime time – Validated
Hypothesis 2: The “virtual model” works — aka the lab-less discovery biotech
Though Nimbus was formed to be an in silico-driven discovery house, we still could have had our own wet labs, if it were not for another founding hypothesis – that we could perform even better without. This was partly an imperative of the times, coming out of the Great Recession, when startups were pushed to be even more capital efficient with their limited cash. So Nimbus decided, in lieu of operating its own labs, to rely on CROs and academics to conduct all of our experimentation. As the vast majority of Nimbi are chemists, biologists, and other scientists who have worked in labs all their careers, this required a different mindset, but one that quickly became just as fulfilling. As our network of collaborators grew, we were fond of saying “the sun never sets” on work at Nimbus. As the Cambridge teams battle the commute home at night, groups in California, then China, India, Poland, and the UK toil, so that everyone’s favorite time at Nimbus is the morning, when fresh data arrive!
The Nimbus culture prizes collaborative relationship skills given this model. Honed by working with our founding partner Schrödinger, Nimbus scientists have developed deep working relationships with many groups and have benefited from the great talent exodus from big pharma. I discussed some of the implications of operating in this “biotech gig economy” here. We further innovated the relationship with our partners two years ago, when we began a true risk-sharing integrated discovery services partnership with Charles River Labs (CRL). We had worked with CRL for years, but now we were coupling all of their keen scientific insight and full suite of discovery wet lab services with our scientists and Schrödinger team members. The highly sought-after program this group has been working on has rocketed forward and is set to begin IND-enabling work next year.
Another challenge with the “virtual model” is how to process and handle all the data coming into Nimbus from literally hundreds of different sources, and somehow processing that data, quality checking it, and depositing it securely into a seamless architecture. Rebecca Carazza on the Nimbus team has done yeoman’s work in creating a seamless back office operation, which she recently shared on the BIO IT World stage. The additional burden of logistics, shipping, and work group coordination is not an easy thing to build corporate proficiency around. In fact, we view the years of hard work to get this system right as a core asset of the company and, frankly, a barrier to entry for others.
Hypothesis 2: The “virtual model” works – Validated
Hypothesis 3: The biotech LLC
The year was 2008, Bear Stearns went belly-up, the global financial markets were in free fall, and the first ideas around Nimbus were being pitched. Tough times, quickly forgotten today, but it was back then that the future of venture investing into risky assets like biotech was not so certain. From those problems came an innovative solution: the “LLC model” structure as a means to more efficiently meter investments into specific projects and avoid the double taxation of traditional corporations.
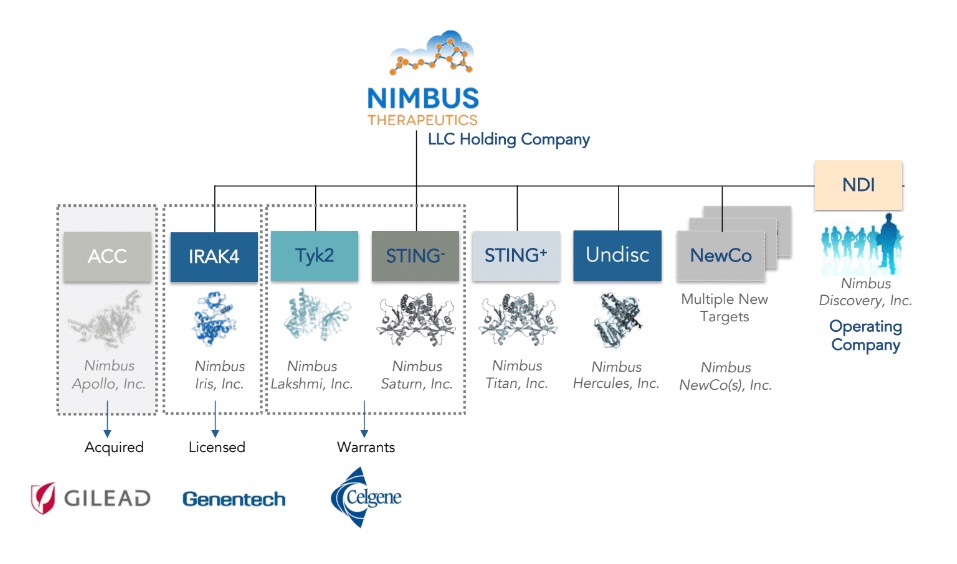
Nimbus was an ideal biotech to introduce the LLC model: our programs are individual collections of intellectual property that revolve around entirely discrete areas of chemical space, ideal for nestling into the various C-corporation subsidiaries under Nimbus Therapeutics LLC. Each of these subsidiaries could then be served by our operating company, Nimbus Discovery, Inc. (or “NDI” as we call it), which in turn would contract master agreements with the hundreds of lab operators described above. The value of this structure has really been twofold. First, for investors the structure provides liquidity when a subsidiary is sold, as it did in 2016 in the sale of Nimbus Apollo, Inc., to Gilead. As I shared at the time, the LLC model worked brilliantly in the case of success. The second value has come in attracting capital to fund our science. In particular, the structure allows great flexibility to partner programs in various deal structures, as illustrated below in three different deal constructs: a sale to Gilead, a license to Genentech, and warrants to Celgene.
For investors, we have often gotten the request to invest in specific subsidiaries, and to date we have eschewed that path, preferring instead to have all investor capital come in at the top LLC level. That choice has allowed alignment of investor interests as we progress a half dozen programs at any given time in our portfolio. Had we had a different cap table for one subsidiary versus another, managing both together, like some chaotic storm of entropy, would have fractured the architecture.
Coupling partnering and capital raising, Nimbus has brought in ~$800 million of capital since inception – $102m in the form of equity, the rest from BD activity. We have spent $155m in R&D so far and have runway to return to Phase 2 clinical efforts with our Tyk2 program. We have returned $570m of capital to investors during this time as well. But we are not done – there are downstream milestones and high-value programmed exits as well as an undisclosed pipeline of new programs that we will begin introducing to the world in the coming months. While we have often thought about the benefits of expanded capital access possible through an IPO, we have so far demurred. As we return to the clinic with our Tyk2 program, we will be revisiting that calculus, as I describe here.
Hypothesis 3: The biotech LLC – Validated
Inspiring Others
Validation of all three originating hypotheses underpinning Nimbus allowed us to contribute to the grander objective of changing how drug discovery and early development can work for small molecules: if imitation is the sincerest form of flattery, Nimbus should be blushing.
We are incredibly proud to see other in silico companies join us in the quest to find the next generation of important medicines — e.g., Morphic using structural biology insights to unlock integrins, or Relay working with the Anton super computer to tackle difficult protein motion problems. Schrödinger itself, which has kept its software approaches at the cutting edge, has introduced much more depth in its own internal drug discovery capabilities. We often get asked if this presents competition for Nimbus; it does not, owing to the formative fact of the extensive exclusivity Nimbus possesses on its pipeline and drug discovery efforts with Schrödinger and other partners. So truly, the more the merrier, and the better for patients and our industry that the novel approaches Nimbus pioneered are gaining traction elsewhere.
After a decade we also have developed an exemplary set of accomplished alumni, who do Nimbus proud. They have gone on to found new companies like HotSpot, to become entrepreneurs in residence at places like GV, and to bring their talents and learnings from Nimbus to other dedicated groups.
Nimbus’ Next Decade
Where will Nimbus be in the next 10 years? The scientific community got a preview of where Nimbus is going last month when, with collaborators at Columbia University and Schrödinger, we published in Nature on the first cryo-EM structure of ATP citrate lyase, with novel Nimbus compounds bound allosterically. Nimbus has rapidly expanded its partnership of protein visualization partners, including deep relationships with crystallization experts at Proteros and elsewhere, and has augmented those with high technology academic centers like Liang Tong’s lab at Columbia, referenced above, as well as other academic cryo-EM centers. Nimbus has pushed even further to explore the select application of protein homology modeling with Cyrus Biotechnology, the UW Baker Lab group bringing Rosetta to commercial application.
We will be doubling down on innovations that couple structure-based drug discovery techniques, such as cryo-EM and other advanced crystallization techniques, with computational methods, including pioneering work we are doing with Schrödinger on ADME profile predictions. One such piece of software developed for Nimbus’ Tyk2 program includes efflux model predictions, a challenge that our team needed to overcome during lead optimization. Other approaches we are pioneering include coupling ligand-based methods with Schrödinger’s FEP+ software to more rapidly iterate and expand chemical space around a target of interest. Our new Chief Scientific Officer, Peter Tummino, formerly led global lead discovery at Janssen and is now busy at work growing both our team and pipeline to equip us for this next decade of designing breakthroughs for patients.
Here’s to 10 remarkable years — and to the next 10 ahead. Cheers!